The world of microorganisms is not quite different from that of humans. Just as the human race is divided into race, religion, social classes, etc., so also is the microbial world divided into different kingdoms, classes, families, groups, characteristics, etc.
The bacterial world is divided mainly into what is called Gram-positives and Gram-negatives based on the colours presented by the cells after staining with some reagents in a process called Gram-staining. Gram staining is a differential stain named after Hans Christian Gram, a Danish bacteriologist who developed the technique in 1884. By using the staining method of Hans Gram, bacteria are classified into two groups based on the physical and chemical properties of their individual cell walls.
Join our WhatsApp ChannelGram-positive bacteria are stained purple or blue when viewed under the immersion objective lens of a light microscope. This is because they have thicker cell walls, which retain the colour of the primary stain called crystal violet. Gram-negatives on the other hand, have thinner cell walls and as such are unable to retain crystal violet when decolourised with ethanol or acetone. This means that when they are treated with the counter stain called safranin or fuchsin, they take up the stain and are coloured pink or red.
READ ALSO: Gut Bacteria Could Checkmate Cardiovascular Diseases By Reducing Cholesterol Levels In Blood
While Gram staining is the first diagnostic tool for identifying a bacterium isolated from a clinical sample, bacteria are not always neatly classified into Gram-positives and Gram-negatives. This is because, there are some bacteria which give indeterminate colours and are classified as Gram-indeterminate or Gram-variable.
Gram-positive bacteria are easier to target with antibiotics even though they have thicker cell walls while infections by Gram-negatives are more difficult to treat with antibiotics despite the bacteria having thinner cell walls. The fundamental reason for this anomaly is that Gram-positive bacterial cell walls are composed of only one structure called peptidoglycan. This structure is layered in such a way as to provide rigidity to the cell walls. While the cell walls of Gram-negative bacteria are composed of the same peptidoglycan, though thinner compared with their Gram-positive counterparts, but in addition, their cell walls are surrounded by an outer membrane that contain lipopolysaccharide (LPS). Anchoring the LPS to the peptidoglycan in Gram-negative bacteria is another structure called lipoprotein. The LPS prevents antibiotics from permeating into the cells of Gram-negative bacteria and given the multilayered architecture of the cell walls of Gram-negative bacteria, diseases caused by the bacteria are difficult to treat.
The gut microbiota consists of all the microorganisms living in the gastrointestinal system. Most of these mixed microorganisms are important to the wellbeing of the body. This means that any disturbance to the gut microbiome equilibrium has unwelcome consequences on the body. Thus, the use of broad-spectrum antibiotics (ie antibiotics that indiscriminately target both Gram-positive and Gram-negative bacteria), often cause a disruption of the gut microbiome equilibrium, giving rise to secondary infections like diarrhoea caused by Clostridioides difficile (formerly known as Clostridium difficile).
There are few, if at all there are, any antibiotics that can specifically target Gram-negative bacteria without also targeting beneficial Gram-positive bacteria in the gut. But that’s about to change if a new antibiotic engineered by Paul Hergenrother and coworkers and proven to work in mice, could also work in humans. And another caveat, if the bacteria do not develop immunity to the antibiotic given their notoriety in that direction.
Paul Hergenrother and coworkers reported in the 29 May 2024 edition of the journal, Nature, the creation of a new antibiotic that specifically targets Gram-negative bacteria without affecting Gram-positives. In a nutshell, the scientists targeted an essential protein exclusively found in Gram-negative bacteria.
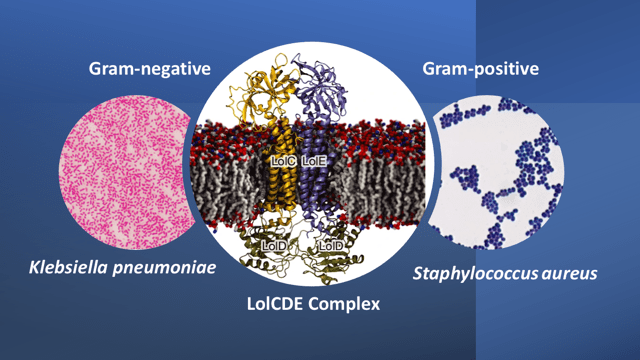
In the Gram-negative bacteria, the transport of lipoprotein between the inner and outer membranes is conducted by a 5-component protein system called the Lol system. This system is absent in Gram-positive bacteria where the lipoprotein is anchored to the cytoplasmic membrane. The beauty of selecting the lipoprotein transport system for design of a selective antibiotic targeting Gram-negatives, is that, it is also conserved across many of the Gram-negative bacteria that make up the ESKAPE pathogens (i.e. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species; only the first two are Gram-positives while the rest are Gram negatives). The ESKAPE pathogens are designated as high-priority pathogens because of their high virulence and frequency of developing antibiotic resistance.
The Lol system is comprised of 5 proteins: LolA is a periplasmic chaperone, LolB is an outer membrane receptor, and LolC, D, and E, together form a complex called the LolCDE complex. This complex is responsible for transporting lipoproteins from the inner membrane to the outer membrane. Armed with this information, the scientists screened many inhibitors against the LolCDE complex with the proviso that the ideal candidate molecules must have high efficacy, high solubility, and the frequency of occurrence of microbial resistance to the molecules must be very low. By this set of criteria and after enhancing some desired properties, Lolamicin (apparently named after the Lol transport system), was identified as a potential candidate molecule.
READ ALSO: Gene Therapy Technology For Sickle-cell Disease Treatment
Mice were exposed to antibiotic-resistant Gram-negative bacteria so that they developed bloodstream infections, and then challenged with lolamicin. All the lolamicin-treated mice survived whereas 87% of the untreated mice died within 3 days of infection.
The mode of action of lolamicin is through competitive inhibition of lipoprotein transport. By physically occupying the lipoprotein-binding site, lolamicin selectively kills pathogenic Gram-negative bacteria, including Escherichia coli, Klebsiella pneumoniae and Enterobacter cloacae. Extensive tests also show that lolamicin is effective against multidrug-resistant clinical isolates of E. coli, K. pneumoniae, and En. cloaca, while having no effect on Gram-positive bacteria or on non-pathogenic Gram-negative commensal bacteria. The reason for this is that lolamicin leverages on the high sequence similarity of the LolCDE complex between E. coli, K. pneumoniae and En. cloacae, as against low sequence similarity between the other bacterial species.
Clostridioides difficile is an opportunistic pathogen that takes advantage of low abundance of other bacteria following a course of antibiotic treatment to proliferate and cause diarrhoea. Due to the specificity of lolamicin, the gut microbiome equilibrium is not affected during tests conducted in mice. Consequently, diarrhoea caused by C. difficile colonisation is prevented. Whereas antibiotics like clindamycin (specific for Gram-positive bacteria) or amoxicillin (a broad-spectrum antibiotic), exerted a severe disruption of the mice gut microbiome so that C. difficile proliferate and cause diarrhoea.
The development of bacteria-specific antibiotics as exemplified by lolamicin in this study, is important in minimising unintended damage to the gut microbiome and preserving patients’ health. However, although the synthetic antibiotic showed promising potentials in laboratory tests with mice, it is unlikely that these results would translate into human use any time soon as it takes over 20 years to develop a candidate antibiotic. The light at the end of the tunnel, however, is the demonstration of proof of concept this work represents.

















Follow Us